FEATURED
GeneSetCart
GeneSetCart is an interactive web-based application that enables users to fetch gene sets from various Common Fund programs data sources, augment these sets with gene-gene co-expression correlations or protein-protein interactions, perform set operations such as union, consensus, and intersection on multiple sets, visualize and analyze these gene sets in a single session.
FEATURED
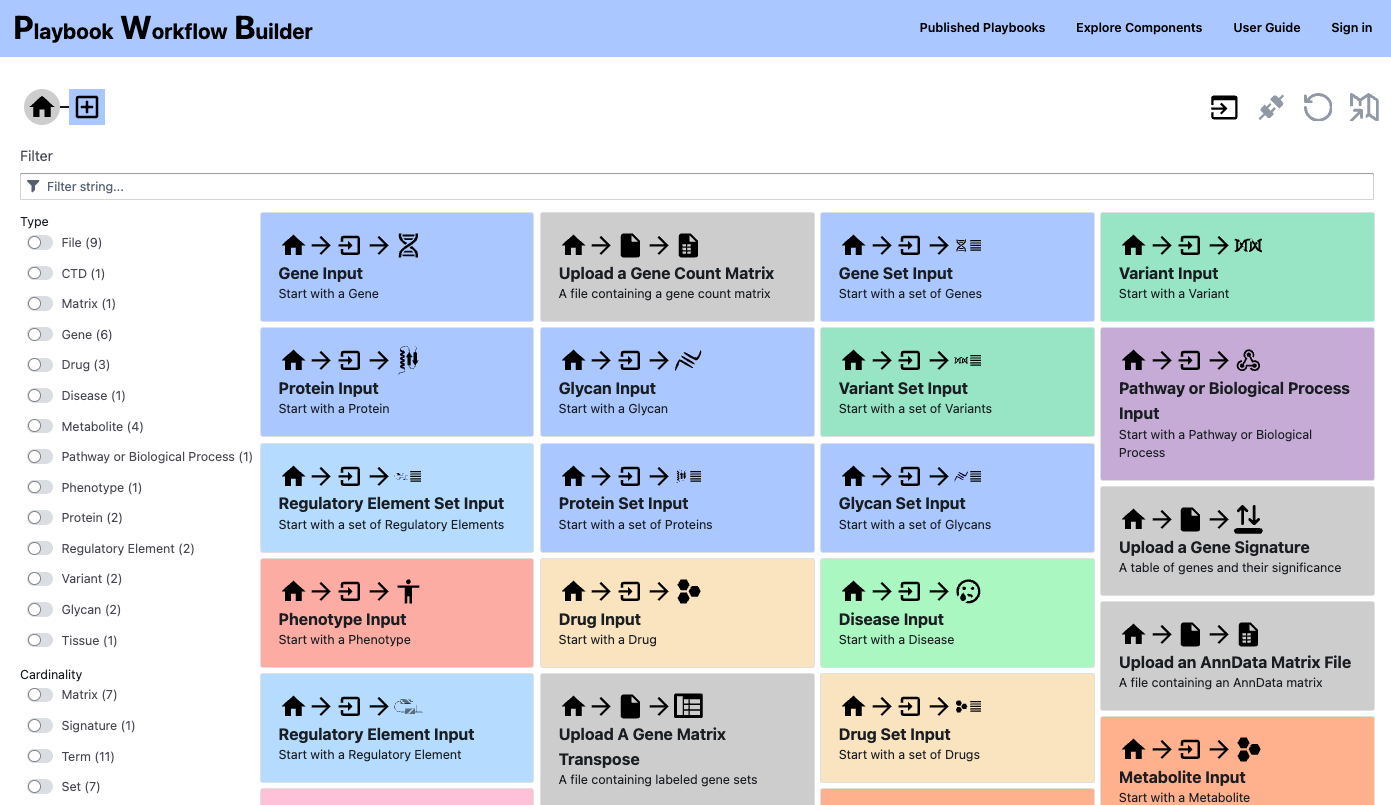
Playbook Workflow Builder
The Playbook Workflow Builder (PWB) is a web-based platform that facilitates knowledge resolution by enabling users to traverse an ever-growing network of input datasets, semantically annotated API endpoints, and data visualization tools. Via a user-friendly user-interface, workflows can be constructed from these building-blocks without technical expertise. The output of these workflows is provided as reports containing textual descriptions, interactive figures, and downloadable tables.
FEATURED
GeneSetCart
GeneSetCart is an interactive web-based application that enables users to fetch gene sets from various Common Fund programs data sources, augment these sets with gene-gene co-expression correlations or protein-protein interactions, perform set operations such as union, consensus, and intersection on multiple sets, visualize and analyze these gene sets in a single session.
FEATURED
Playbook Workflow Builder
The Playbook Workflow Builder (PWB) is a web-based platform that facilitates knowledge resolution by enabling users to traverse an ever-growing network of input datasets, semantically annotated API endpoints, and data visualization tools. Via a user-friendly user-interface, workflows can be constructed from these building-blocks without technical expertise. The output of these workflows is provided as reports containing textual descriptions, interactive figures, and downloadable tables.
FEATURED
GeneSetCart
GeneSetCart is an interactive web-based application that enables users to fetch gene sets from various Common Fund programs data sources, augment these sets with gene-gene co-expression correlations or protein-protein interactions, perform set operations such as union, consensus, and intersection on multiple sets, visualize and analyze these gene sets in a single session.
FEATURED
Playbook Workflow Builder
The Playbook Workflow Builder (PWB) is a web-based platform that facilitates knowledge resolution by enabling users to traverse an ever-growing network of input datasets, semantically annotated API endpoints, and data visualization tools. Via a user-friendly user-interface, workflows can be constructed from these building-blocks without technical expertise. The output of these workflows is provided as reports containing textual descriptions, interactive figures, and downloadable tables.
All Tools and Workflows
Explore different tools and workflows that utilizes data from different CFDE participating programs
@CFDE Workbench 2026
The CFDE Workbench is actively being developed and maintained by the CFDE Data Resource Center (DRC).The DRC is funded by OT2OD036435 from the Common Fund at the National Institutes of Health.
The CFDE Workbench is actively being developed and maintained by the CFDE Data Resource Center (DRC).The DRC is funded by OT2OD036435 from the Common Fund at the National Institutes of Health.
@CFDE Workbench 2026